IRDye® 700 Phosphoramidite
Components
4200-33 | IRDye 700 Phosphoramidite, 100 μmol |
Product Description
IRDye 700 phosphoramidite is a near-infrared fluorescent dye used to label DNA molecules prepared in a DNA synthesis machine. The dye is attached to the 5' end of the protected, support-bonded oligonucleotide via fast deprotection (Expedite™) phosphoramidite chemistry.
On syntheses at 200 nmol scale, typical crude yields of dye-labeled oligonucleotides are 100 to 150 nmol. Standard phosphoramidites and deprotection conditions can also be used.
This protocol was developed on a PerSeptive Biosystems 8909 DNA synthesizer and may require optimization for use with other systems.
Properties
Absorption Max (λmax, methanol): | 680 nm |
| Absorption Max (aqueous oligonucleotide): | 685 nm |
| Emission Max: | 705 nm |
| Molar Absorptivity: | 170,000 |
Structure
Chemical Formula: | C52H67N4O5PS |
| Molecular Weight: | 891.16 g/mol |
| Exact Mass: | 890.46 |
Required Equipment
This product requires the LICORbio Model 4300 or dual-laser IR2 Model 4200 DNA Analyzer. Oligonucleotides labeled with IRDye 700 dye are not compatible with LICORbio Model 4000 Series or single dye Model 4200 DNA Sequencers.
PerSeptive Biosystems 8909 DNA Synthesizer or equivalent
CPG column, 500Å, 200 nanomole (PerSeptive Biosystems Expedite™: GEN084167, GEN084168, GEN084169, GEN062680; or Standard: GEN062650, GEN062660, GEN062670, GEN062680 or equivalent)
A, C, G, and T phosphoramidites (PerSeptive Biosystems Expedite™: GEN084282 or Standard: GEN067040 or equivalent)
Microcentrifuge, standard and filter tubes
Gradient HPLC system, reverse phase C18, for product analysis and purification (if desired)
Required Reagents
IRDye 700 Phosphoramidite (P/N 4200-33)
Synthesis reagents
Oxidizer solution
Capping reagent solution
Deblocking reagent solution
Activator solution
Acetonitrile (PerSeptive Biosystems Expedite™: GEN084800 or Standard: GEN089800 or equivalent)
Ammonia
HPLC reagents
HPLC solvents
Buffer
Acetonitrile (HPLC grade)
Triethylammonium acetate
Precautions
Handling Phosphoramidites
All phosphoramidites are sensitive to moisture and oxygen. The handling procedures described here are good practice in general. You should not need to modify your standard procedures for similar products when using IRDye 700 Phosphoramidite.
Protect the product from air and moisture, and only use dry, oxygen-free, DNA synthesis grade acetonitrile to prepare the amidite solutions.
Solutions
Oxidizer
The oxidizer solution supplied by PerSeptive Biosystems (GEN089850 or equivalent) should be used. The recipe for an equivalent oxidizer solution is: 4.3 g iodine, 90 ml water, 4 ml pyridine, and 900 ml THF.
Harsher oxidizer solutions supplied by some manufacturers, such as ABI, are not recommended.
Acetonitrile
Acetonitrile solutions of IRDye 700 can be left on the instrument for up to two weeks. However, if it will be more than a few days to the next synthesis, we recommend refrigerating the solution. There is some loss of coupling efficiency of the solutions on standing, but they maintain reasonable efficiency after storage for two weeks in dry, oxygen-free solution.
Flushing the lines of the synthesizer with acetonitrile (or methanol followed by acetonitrile) is generally sufficient to remove any traces of dye.
Ammonia
The ammonia solution used in the deprotection of the oligonucletides should be handled in a fumehood.
Protocol
Step 1. Prepare IRDye 700 Phosphoramidite Solution
Make a 0.1 M solution of IRDye 700 in acetonitrile by adding 1.00 ml of acetonitrile to the 100 μmol package of IRDye 700 Phosphoramidite.
Choose a clean, dry syringe and needle of appropriate size.
Pierce the seal of the acetonitrile bottle with the needle and slowly withdraw slightly more than 1.00 ml of acetonitrile.
A good practice is to keep a source of dry nitrogen or argon gas to displace the acetonitrile that will be removed from the bottle. A needle connected to a stream of the gas and a bubbler is a convenient way to do this. Another approach is to use a second syringe barrel filled with desiccant for the vent. However, this approach does not remove the oxygen. Use a similar setup to allow for the escape of the gas displaced when the acetonitrile is added to the phosphoramidite vial.
Perform steps 3 to 5 rapidly to minimize exposure of the acetonitrile to air.
Remove the syringe and needle from the acetonitrile bottle.
Invert the syringe and force any air bubbles out the needle. Push the excess acetonitrile from the syringe into an appropriate waste container.
Inject the acetonitrile into the sealed IRDye 700 Phosphoramidite vial.
Remove the needle and allow the IRDye 700 Phosphoramidite to dissolve.
Swirl the solution occasionally until no undissolved particles are observed on the sides of the vial.
Step 2. Perform DNA Synthesis Program
The basic PerSeptive Biosystems DNA synthesis protocol (“DNA 0.2 μmol”) is used until the last step involving the dye amidite. The commands used for the dye coupling on the PerSeptive 8909 are shown in the table.
A total volume of approximately 80 μl of 0.1 M IRDye 700 phosphoramidite solution (8 μmol, five 16 μl pulses) is used per labeling reaction. The instrument should be used in the “Final DMT On” mode to avoid unnecessary exposure of the dye to the detritylation reagent. The IRDye 700 phosphoramidite does not contain any trityl group. Position 7 is used for the IRDye 700 phosphoramidite solution.
| Function Coupling | Mode | Amount /Arg1 | Time (sec) /Arg2 | Description |
| 1/*Wsh | */PULSE | 5 | 0 | Flush system with Wsh |
| 2/*Act | */PULSE | 5 | 0 | Activator to column |
| 24/*7 + Act | */PULSE | 5 | 0 | Monomer + Act to column |
| 2/*Act | */PULSE | 1 | 0 | Chase with Act |
| 2/*Act | */PULSE | 4 | 63 | Couple monomer |
| 0/*Default | */WAIT | 0 | 180 | 3 min wait |
| 1/*Wsh | */PULSE | 2 | 31 | Couple monomer |
| 0/*Default | */WAIT | 0 | 180 | 3 min wait |
| 1/*Wsh | */PULSE | 13 | 0 | Flush system with Wsh |
Step 3. Deprotect Primer
Place the CPG beads from the column into a 1.5 ml microcentrifuge tube.
Add 800 μl of concentrated ammonium hydroxide solution to the tube.
Close the tube and mix the contents thoroughly on a vortex mixer.
If you are using the Expedite chemistry, allow the tube to stand in the dark at room temperature for 2 hours. For standard deprotection chemistry, let the tube stand for 16 hours.
Mix the solution thoroughly on a vortex mixer, then spin the tube briefly in a microcentrifuge.
Decant the solution from the beads into another 1.5 ml microcentrifuge tube using a pipet.
Strip off the aqueous ammonia in a vacuum drying unit.
Re-suspend the dry pellet in 100 μl of sterile, ultrapure water.
Load the primer suspension onto a sterile, labeled acetate microfuge filter (1.5 ml, 0.22 μm).
Wash the pellet tube with another 25 μl of sterile, ultrapure water, and transfer the wash to the filter.
Spin the filter tube at 7500 x g for 5 minutes. Remove the filter and close the tube containing the filtered primer solution.
Step 4. Measure Crude Synthesis Yield (optional)
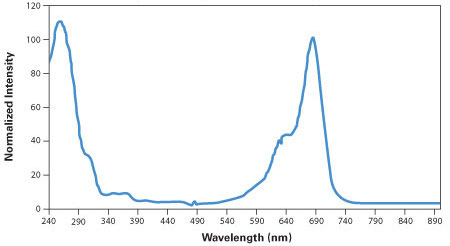
Dilute 1.0 μL of the filtered primer solution with 399.0 μL of sterile ultrapure water.
Obtain the UV/Vis absorption spectrum of the resulting solution (see Figure 40).
Calculate the crude yield from the absorbance value at 685 nm (A685), the absorptivity (170 μL nmol-1 cm-1), the path length (b cm), the dilution factor (400), and the total primer solution volume (120 μL):
For example, a measured absorbance of 0.59 in a 1 cm cell would correspond to a crude yield of 167 nmol.
Step 5. Evaluate Primer Purity
The crude oligonucleotides are sufficiently pure for many purposes. Their purities can be checked with analytical HPLC. Preparative HPLC can be used when the highest purity is needed.
It is convenient to monitor at least two wavelengths with the HPLC detector. Detection at 680 nm shows just the dye-labeled components in the sample, while 260 or 280 nm will also reveal unlabeled products, such as the failure sequences of the synthesis. These HPLC conditions can be used for analysis and purification.
Solvents: | A = 0.1 M triethylammonium acetate in 4% acetonitrile/96% water B = 0.1 M triethylammonium acetate in 80% acetonitrile/20% water |
| Column: | C18, 5 μm, 300 Å, 10 × 100 mm |
| Flow: | 1.7 ml/min |
| Detection: | 280 and 680 nm |
| Gradient: | 10–45% B over 5 minutes 45–100% B over 15 minutes 100% B for 5 minutes |
Analytical HPLC
Typical chromatograms for both wavelengths are shown in Figure 41 for an oligonucleotide prepared with Expedite™ chemistry. The main peak is the desired product. Synthesis failure sequences are more hydrophilic, so these components elute earlier (2–10 min).
Dilute 1 μl of the filtered, deprotected oligonucleotide solution from step 3 with 99 μl of sterile, pure water.
Inject 10 μl of the diluted solution for the analytical run.
Preparative HPLC
If LC purification is desired, all or part of the filtered, deprotected oligonucleotide solution from step 3 is directly injected. Collect appropriate fractions and remove the solvents and the triethylammonium acetate buffer in a vacuum drying unit.
Prepare HPLC Solution
Prepare the oligonucleotide in 2 mM EDTA solution. For a 1.0 μM (1.0 pmol/μL) solution, dilute the dye-labeled oligonucleotide to obtain 0.17 absorbance units (685 nm) in a 1 cm cell.

